KavaTasteGood
Kava Curious
As I stated before, I’m a young doctor specialized in molecular modeling. In this post, we will explore how molecular docking can be used to search for the binding site of kavalactones that are known to interact with the GABA-A receptor in the brain [1] but have no specific binding site discovered yet. By simulating the interaction between kavalactones and the receptor, molecular docking can help to identify the most likely binding site and orientation of the compounds.
1. The GABAergic transmission
GABAergic and glutamatergic neurotransmissions are crucial for brain function. GABA inhibits/slow down neuronal activity and is important for relaxation, sleep, and anxiety regulation. Glutamate excites/speed up neuronal activity and is important for learning, memory, and cognitive processing. The balance between the two is essential for brain health; an imbalance can lead to neurological and psychiatric disorders such as epilepsy, anxiety, depression, and schizophrenia.
There are two types of GABA receptors in the central nervous system (figure 1): GABA-A and GABA-B receptors. GABA-A receptors are ionotropic receptors that directly control the flow of ions into and out of the cell and are responsible for the majority of fast inhibitory neurotransmission. GABA-B receptors are metabotropic receptors that indirectly control ion channels through intracellular signaling pathways and are responsible for slower and more prolonged inhibitory neurotransmission. The different properties and distributions of these receptors allow them to play distinct roles in the regulation of brain activity and the maintenance of overall neurological health. Here, we will focus on the GABA-A receptor.
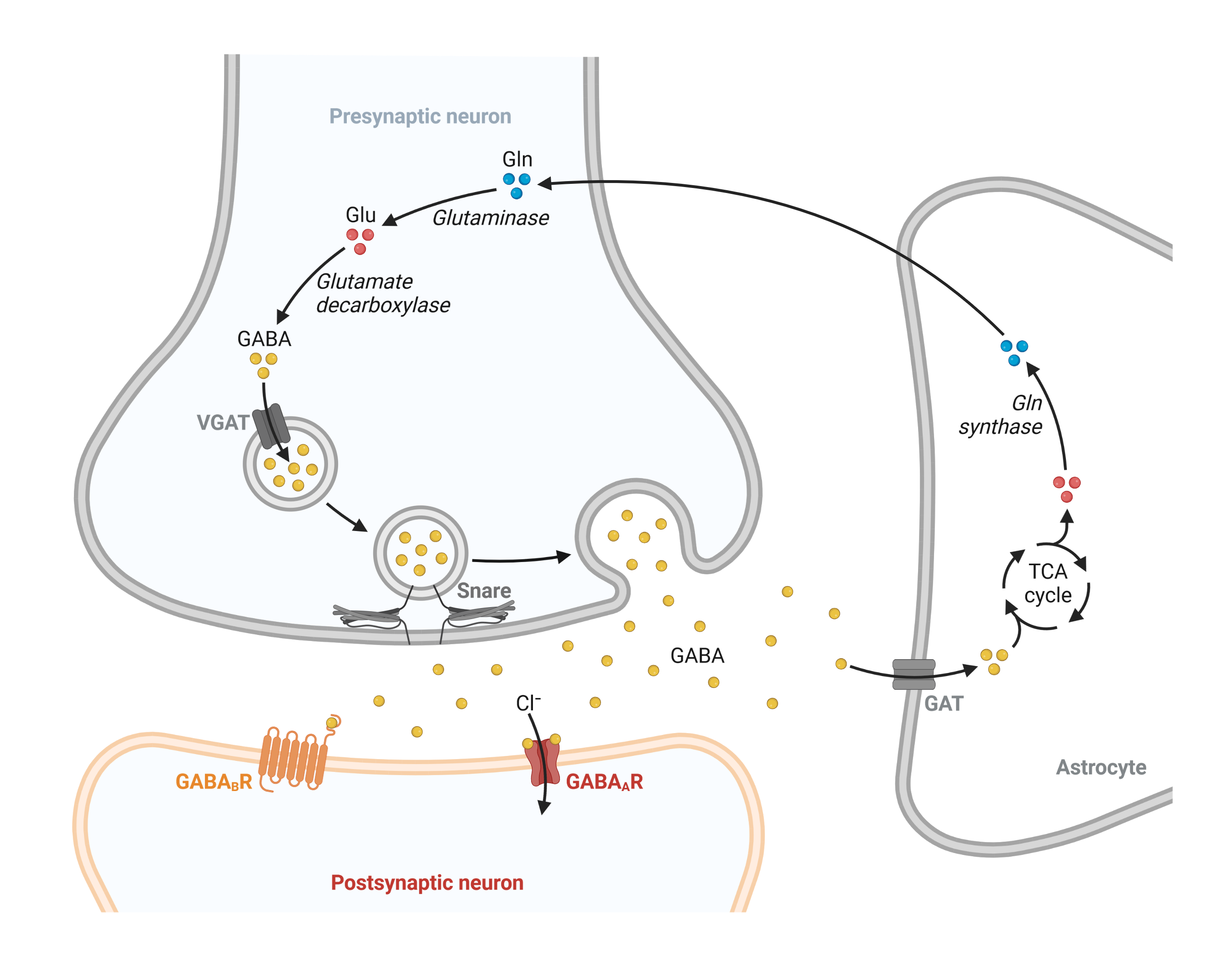
Figure 1: Overview of the GABA cycle.
2. The GABA-A receptor
The GABA-A receptor is composed of five protein subunits that come together to form a functional receptor complex. These subunits can be drawn from a variety of different gene families and can combine in many different ways to form a diverse array of receptor subtypes. In general, the receptor complex contains two alpha subunits, two beta subunits, and one gamma subunit, although there can be some variability in the exact subunit composition depending on the specific subtype of the receptor (figure 2).
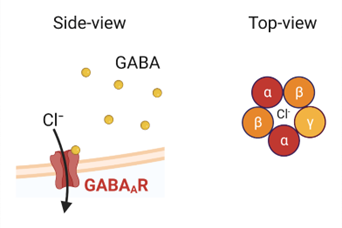
Figure 2: The GABA-A receptor is a pentameric ion channel.
Here I will work on an experimental structure of the GABA-A receptor that was published in 2018 [2] (pdb: 6D6U), which was obtained using a cutting-edge imaging technique called cryo-electron microscopy (cryo-EM). This technique allows researchers to visualize the structure of large macromolecules, such as receptors, at high resolution by freezing them in a thin layer of ice and imaging them with an electron microscope. By having access to the coordinates of the atoms in the molecule, it is possible to use visualization software to directly observe the protein structure, as shown in Figure 3.
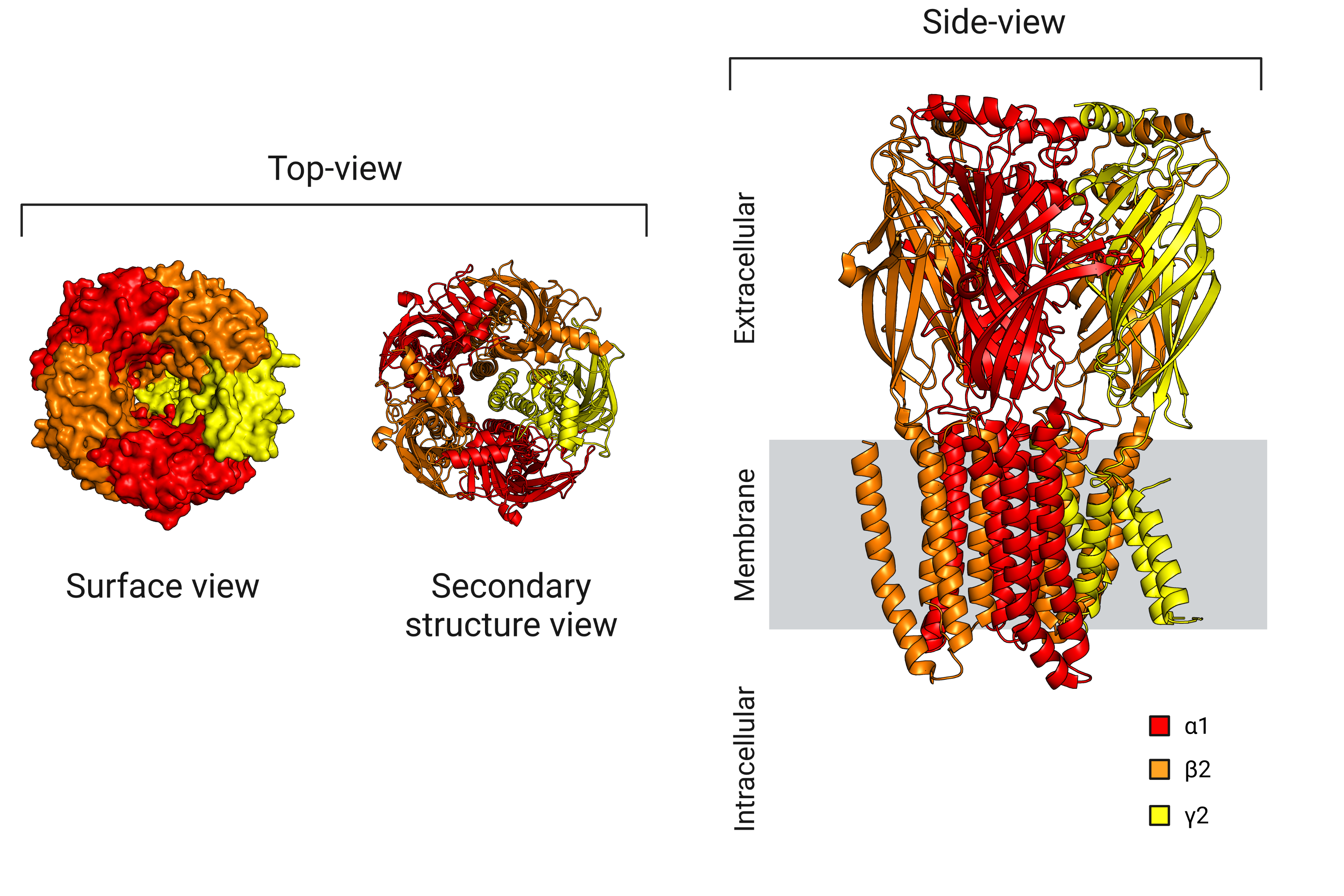
Figure 3: Visualization of the GABA-A receptor structure.
The structure was generated in the presence of both GABA and flumazenil, which provides an opportunity to identify the specific locations of the two orthosteric binding sites (where GABA binds to activate the ion channel) and the benzodiazepine allosteric binding site (where flumazenil binds to facilitate the opening of the channel by GABA), shown in figure 4.
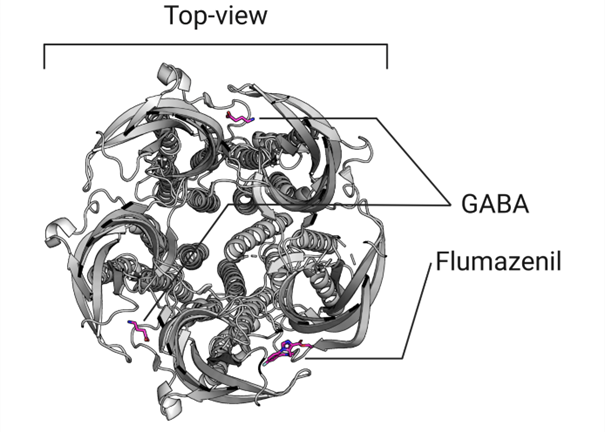
Figure 4: Top-view of the orthosteric and allosteric site of the GABA-A receptor.
Flumazenil is a benzodiazepine antagonist that blocks the activity of benzodiazepines by competing for their binding site. Interestingly, the inhibitory effect of kavalactones on the GABA-A receptor is not affected by flumazenil, indicating that if kavalactones do bind to the receptor, they must do so at a different site. The question then becomes: where exactly do kavalactones bind to the GABA-A receptor if not at the benzodiazepine site or the orthosteric site?
3. Molecular docking
To identify the binding sites of the six kavalactones that show GABAergic activity on the GABA-A receptor, I plan to use blind molecular docking simulations. For this, I will employ a novel docking approach that uses deep learning algorithms to improve the accuracy and efficiency of the simulations, allowing for more precise predictions of the binding locations and orientations of the kavalactones [3]. Additionally, I will dock GABA and flumazenil as negative controls to confirm that they bind to their known binding sites on the GABA-A receptor.
Figure 5 (left) shows that the molecular docking simulations were able to identify the flumazenil binding pocket on the GABA-A receptor, although the predicted binding pose was inverted. In contrast, Figure 5 (right) demonstrates that the molecular docking simulations successfully identified the correct binding site and orientation of GABA on the GABA-A receptor.
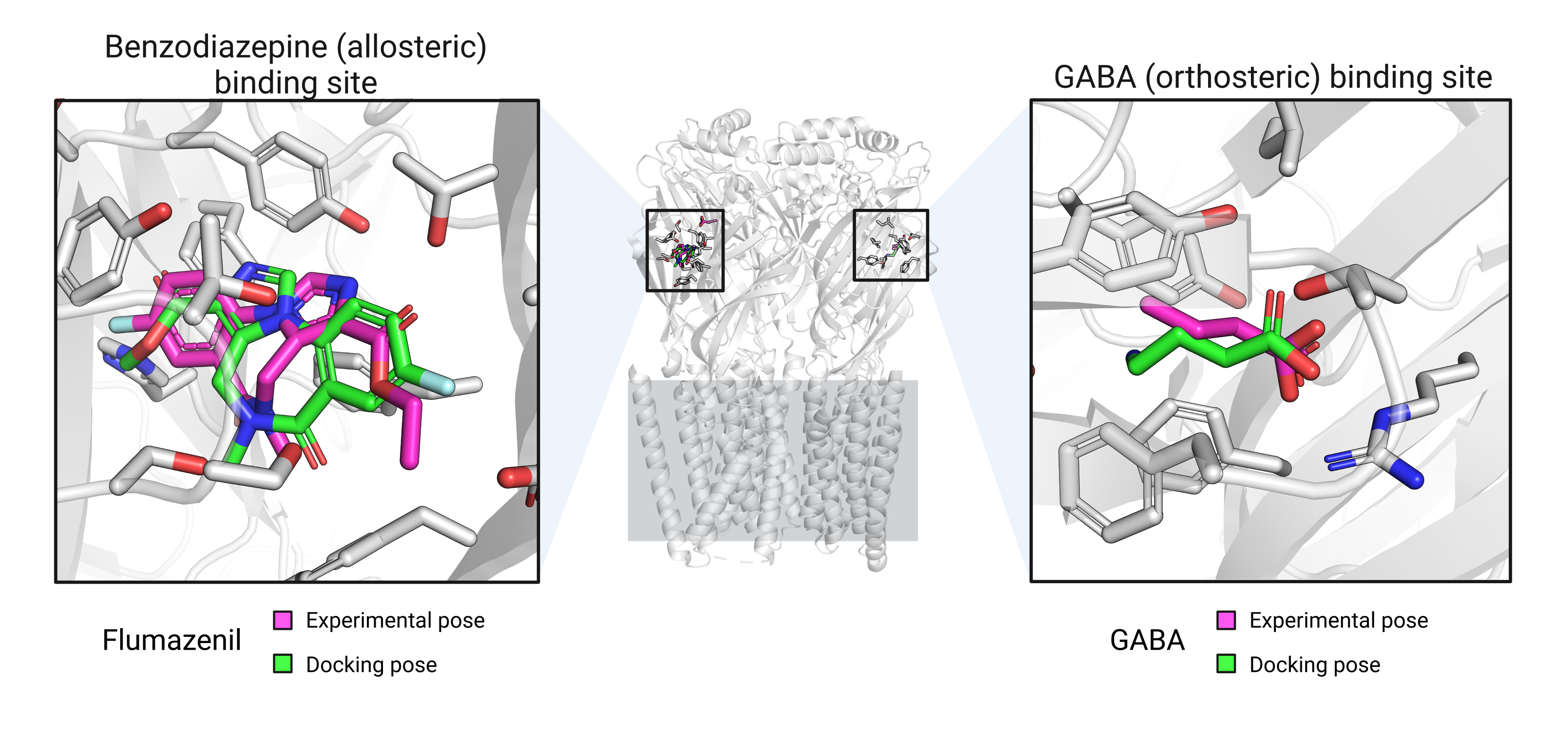
Figure 5: Best docking poses for flumazenil and GABA.
As depicted in Figure 6, the molecular docking simulations revealed that all of the kavalactones docked to the benzodiazepine allosteric binding site on the GABA-A receptor. It is noteworthy that the findings from the molecular docking simulations contradict the experimental evidence that kavain does not bind to the benzodiazepine GABA-A receptor allosteric binding site [4]. Molecular docking simulations have several limitations, such as the inability to consider protein flexibility, solvent effects, and the role of dynamics in ligand binding. These limitations could potentially account for the discrepancy between the molecular docking results and experimental evidence regarding the lack of kavain binding to the GABA-A receptor benzodiazepine allosteric binding site. Therefore, while molecular docking simulations can be a useful tool for identifying putative binding sites and orientations, the results should always be interpreted with caution and corroborated by additional experimental evidence.
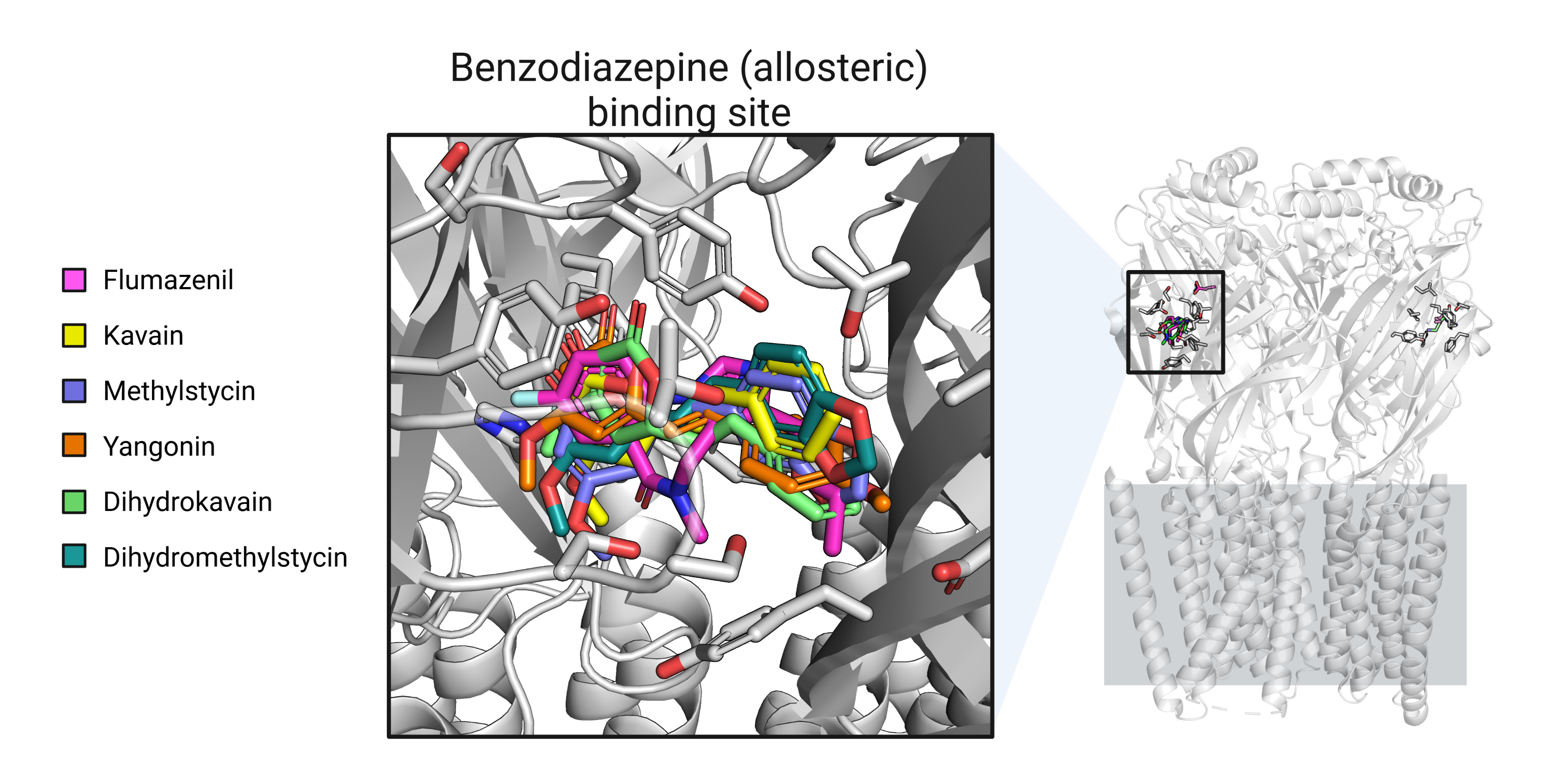
Figure 6: Docking best poses for all kavain analogs.
While the exact location of the GABA-A receptor binding site for kavain and its analogs remains a mystery, I hope you found this information intriguing and thought-provoking. Please feel free to ask me any questions you may have.
1. Singh, Y. N. & Singh, N. N. Therapeutic Potential of Kava in the Treatment of Anxiety Disorders. CNS Drugs 16, 731–743 (2002).
2. Zhu, S. et al. Structure of a human synaptic GABAA receptor. Nature 559, 67–72 (2018).
3. Corso, G., Stärk, H., Jing, B., Barzilay, R. & Jaakkola, T. DiffDock: Diffusion Steps, Twists, and Turns for Molecular Docking. Preprint at https://doi.org/10.48550/arXiv.2210.01776 (2023).
4. Chua, H. C. et al. Kavain, the Major Constituent of the Anxiolytic Kava Extract, Potentiates GABAA Receptors: Functional Characteristics and Molecular Mechanism. PLOS ONE 11, e0157700 (2016).
1. The GABAergic transmission
GABAergic and glutamatergic neurotransmissions are crucial for brain function. GABA inhibits/slow down neuronal activity and is important for relaxation, sleep, and anxiety regulation. Glutamate excites/speed up neuronal activity and is important for learning, memory, and cognitive processing. The balance between the two is essential for brain health; an imbalance can lead to neurological and psychiatric disorders such as epilepsy, anxiety, depression, and schizophrenia.
There are two types of GABA receptors in the central nervous system (figure 1): GABA-A and GABA-B receptors. GABA-A receptors are ionotropic receptors that directly control the flow of ions into and out of the cell and are responsible for the majority of fast inhibitory neurotransmission. GABA-B receptors are metabotropic receptors that indirectly control ion channels through intracellular signaling pathways and are responsible for slower and more prolonged inhibitory neurotransmission. The different properties and distributions of these receptors allow them to play distinct roles in the regulation of brain activity and the maintenance of overall neurological health. Here, we will focus on the GABA-A receptor.
Figure 1: Overview of the GABA cycle.
2. The GABA-A receptor
The GABA-A receptor is composed of five protein subunits that come together to form a functional receptor complex. These subunits can be drawn from a variety of different gene families and can combine in many different ways to form a diverse array of receptor subtypes. In general, the receptor complex contains two alpha subunits, two beta subunits, and one gamma subunit, although there can be some variability in the exact subunit composition depending on the specific subtype of the receptor (figure 2).
Figure 2: The GABA-A receptor is a pentameric ion channel.
Here I will work on an experimental structure of the GABA-A receptor that was published in 2018 [2] (pdb: 6D6U), which was obtained using a cutting-edge imaging technique called cryo-electron microscopy (cryo-EM). This technique allows researchers to visualize the structure of large macromolecules, such as receptors, at high resolution by freezing them in a thin layer of ice and imaging them with an electron microscope. By having access to the coordinates of the atoms in the molecule, it is possible to use visualization software to directly observe the protein structure, as shown in Figure 3.
Figure 3: Visualization of the GABA-A receptor structure.
The structure was generated in the presence of both GABA and flumazenil, which provides an opportunity to identify the specific locations of the two orthosteric binding sites (where GABA binds to activate the ion channel) and the benzodiazepine allosteric binding site (where flumazenil binds to facilitate the opening of the channel by GABA), shown in figure 4.
Figure 4: Top-view of the orthosteric and allosteric site of the GABA-A receptor.
Flumazenil is a benzodiazepine antagonist that blocks the activity of benzodiazepines by competing for their binding site. Interestingly, the inhibitory effect of kavalactones on the GABA-A receptor is not affected by flumazenil, indicating that if kavalactones do bind to the receptor, they must do so at a different site. The question then becomes: where exactly do kavalactones bind to the GABA-A receptor if not at the benzodiazepine site or the orthosteric site?
3. Molecular docking
To identify the binding sites of the six kavalactones that show GABAergic activity on the GABA-A receptor, I plan to use blind molecular docking simulations. For this, I will employ a novel docking approach that uses deep learning algorithms to improve the accuracy and efficiency of the simulations, allowing for more precise predictions of the binding locations and orientations of the kavalactones [3]. Additionally, I will dock GABA and flumazenil as negative controls to confirm that they bind to their known binding sites on the GABA-A receptor.
Figure 5 (left) shows that the molecular docking simulations were able to identify the flumazenil binding pocket on the GABA-A receptor, although the predicted binding pose was inverted. In contrast, Figure 5 (right) demonstrates that the molecular docking simulations successfully identified the correct binding site and orientation of GABA on the GABA-A receptor.
Figure 5: Best docking poses for flumazenil and GABA.
As depicted in Figure 6, the molecular docking simulations revealed that all of the kavalactones docked to the benzodiazepine allosteric binding site on the GABA-A receptor. It is noteworthy that the findings from the molecular docking simulations contradict the experimental evidence that kavain does not bind to the benzodiazepine GABA-A receptor allosteric binding site [4]. Molecular docking simulations have several limitations, such as the inability to consider protein flexibility, solvent effects, and the role of dynamics in ligand binding. These limitations could potentially account for the discrepancy between the molecular docking results and experimental evidence regarding the lack of kavain binding to the GABA-A receptor benzodiazepine allosteric binding site. Therefore, while molecular docking simulations can be a useful tool for identifying putative binding sites and orientations, the results should always be interpreted with caution and corroborated by additional experimental evidence.
Figure 6: Docking best poses for all kavain analogs.
While the exact location of the GABA-A receptor binding site for kavain and its analogs remains a mystery, I hope you found this information intriguing and thought-provoking. Please feel free to ask me any questions you may have.
1. Singh, Y. N. & Singh, N. N. Therapeutic Potential of Kava in the Treatment of Anxiety Disorders. CNS Drugs 16, 731–743 (2002).
2. Zhu, S. et al. Structure of a human synaptic GABAA receptor. Nature 559, 67–72 (2018).
3. Corso, G., Stärk, H., Jing, B., Barzilay, R. & Jaakkola, T. DiffDock: Diffusion Steps, Twists, and Turns for Molecular Docking. Preprint at https://doi.org/10.48550/arXiv.2210.01776 (2023).
4. Chua, H. C. et al. Kavain, the Major Constituent of the Anxiolytic Kava Extract, Potentiates GABAA Receptors: Functional Characteristics and Molecular Mechanism. PLOS ONE 11, e0157700 (2016).
Attachments
-
75.7 KB Views: 44
-
136.4 KB Views: 52
-
138 KB Views: 46
-
150.1 KB Views: 58
Last edited: